Data input (tutorial): Difference between revisions
| Line 76: | Line 76: | ||
To create a network from one of the above mentioned data formats (adjacency matrix, link list, adjacency list) use the [[file menu]], click on '''open...''', select ''files of type'' '''CSV files (.txt, .csv)''' in the file chooser, navigate to the file you want to open, and click on the '''ok''' button. Then the [[import options dialog]] opens, where you select the appropriate data format in the topmost drop-down menu. | To create a network from one of the above mentioned data formats (adjacency matrix, link list, adjacency list) use the [[file menu]], click on '''open...''', select ''files of type'' '''CSV files (.txt, .csv)''' in the file chooser, navigate to the file you want to open, and click on the '''ok''' button. Then the [[import options dialog]] opens, where you select the appropriate data format in the topmost drop-down menu. | ||
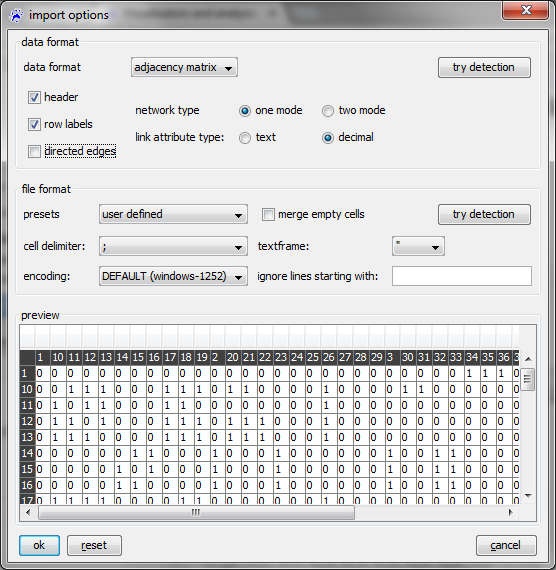
Depending of the data format, the import options dialog shows you the various settings to be made, to interpret the data correctly. | Depending of the data format, the import options dialog shows you the various settings to be made, to interpret the data correctly. The image below shows the import options dialog, when selecting ''adjacency matrix'' as data format. The visualization and analysis tutorial explains the [[Visualization and analysis (tutorial)#Importing networks from adjacency matrix files|import of data from an adjacency matrix in detail]], using an exemplary dataset. A description of importing all (other) data formats is given on the [[import options dialog]] page. Note that the import options dialog is designed to import information about nodes and links, not about attributes. The only exception is the [[Import options dialog#Node list files|import of node lists]], where a network of isolated nodes and their attributes will be created. | ||
[[File:Import_options_dialog_adjacency_matrix.png]] | |||
== Importing node and link attributes == | == Importing node and link attributes == | ||
Revision as of 08:19, 19 May 2015
Normally, visone reads network data from GraphML files, which should never cause any problems. However, in some cases it is necessary to import data stemming from other sources that can, for instance, export adjacency matrices to comma-separated-value (CSV) tables. This tutorial guides you through the various possibilities to input data into visone.
The usual way: read GraphML
GraphML is the usual file format for visone; it encodes the three types of information that are contained in visone networks: network structure, attributes, and graphical information and it is the only format that does so. To read network data from GraphML files use the file menu, click on open..., select files of type .graphml, and click on the ok button.
The other file types are only needed when you want to import data from other sources that cannot output GraphML.
An overview about the other possibilities
Apart from GraphML, visone can read network data from files that are exported by other network analysis software, including UCINET, Pajek, Siena, and some more. Opening these files is also done via the file menu by selecting the appropriate file type. More information about reading these file types is provided in the section about other supported formats.
A more basic option that should be feasible in most situations is to read network data from comma-separated-value (CSV) files. CSV files are plain-text files, for example looking like this
;A;B;C;D A;0;1;1;1 B;1;0;1;0 C;1;1;0;0 D;1;0;0;0
that can be created by spread sheet editors (such as MS Excel), statistical software, most network analysis software, and many more. However, reading data from CSV files is more error-prone since these files do not come with an unequivocal definition about how to interpret them (rather you have to tell the program how these files should be interpreted). Most of this tutorial is dedicated to the import of CSV files.
Personal networks collected with the EgoNet software can be converted to GraphML with the EgoNet2GraphML converter. This is illustrated in the tutorial on personal networks.
Wikipedia edit networks can be imported after converting edit history files with the WikiEvent software. This is illustrated in the tutorial on Wikipedia edit networks.
If nothing else works, you can make use of the visone R console that allows access to the R environment for statistical computing or you can read data via the KNIME connection that connects visone to a comprehensive data mining workflow tool. Both environments, R and KNIME, provide very general and configurable methods for data input and, in addition, enable you to preprocess and/or filter the data with relatively low effort. We provide more information about these possibilities in the section "if nothing else works".
Yet another possibility to create networks directly in visone is to enter them manually (which is only appropriate if the neworks are small and the data is not yet available in electronic format). This option is illustrated in the tutorial introducing the visual network editor.
The variants of comma-separated-value (CSV) files
A comma-separated-value file can be thought of as a plain-text file that encodes a table, i.e., a data array that has rows and columns - sometimes also referred to as a matrix. In a network context, there are different possibilities to encode information about nodes, links, or attribute information in such tables. The first three (adjacency matrices, link lists, and adjacency lists) provide information about nodes and links and can be opend via the file menu and the import options dialog. The fourth table displays an attribute table, that provides information about node or link attributes. Those data files will be imported via the attribute manager. What follows is a short characterization of these file types. A more exhaustive explanation about how to import those data tables in visone is given in the following sections.
Adjacency matrix files. An adjacency matrix encodes for all pairs of nodes (indexing the rows and columns of the table) whether or not there is a link connecting these nodes. An example is shown in the following.
;A;B;C;D A;0;1;1;1 B;0;0;1;0 C;1;1;0;0 D;1;0;0;0
The first row and the first column are the labels of the nodes; the remaining part encodes whether there is a link from the node indexing the row to the node indexing the column. For instance, the character 1 in the row indexed by A and the column indexed by B indicates that there is a link going from A to B; the 0 in the row B and column A indicates that there is no link in the reverse direction.
Link list files. A link list contains as many rows as there are links in the network and (in its most basic form) a link list contains two columns where the entry in the first column is the identifier for the source node and the entry in the second column denotes the target node of the link. The following example
A;C C;B B;A A;D
defines four links: from node A to node C, from C to B, etc. Note that link lists use less space than adjacency matrices - especially if the network is very sparse (i.e., when the number of links divided by the number of node pairs is a small value close to zero).
Adjacency list files. An adjacency list has as many rows as there are nodes in the network. Each row may have a different length and the row associated with a node lists all neighbors of this node. For instance, in the following example
1;2;4;5 2;1;5 3;5 4;1 5;1;2;4
the adjacency list defines for node 1 three (outgoing) links: to node 2, node 4 and node 5. Node 2 has links to node 1 and node 5, etc.
Attribute tables. An attribute table has one row that lists the attribute names (in the example below this is id;age;smokes followed by as many rows as there are nodes in the network.
id;age;smokes A;23;false B;28;true C;19;true D;27;false
One of the columns (id in the example above) lists the unique node identifiers (it is not necessarily the first row and not necessarily labeled id). The other columns list the attribute values of the attribute whose name is given in the respective column header.
Note: visone does not allow to simultaneously input an adjacency matrix together with given attributes in one file. That is, a file like
;A;B;C;D;age;smokes A;0;1;1;1;23;false B;1;0;1;0;28;true C;1;1;0;0;19;true D;1;0;0;0;27;false
(having the interpretation that the first four columns specify an adjacency matrix and the last two columns define node attribute values) cannot be opened. Rather you have to split this into two separate files, one containing the adjacency matrix which can be opened via the file menu and the other containing the attribute table which can be opened via the attribute manager. (For instance, in MS Excel you could split the file by selecting columns, copy them, and past them into a new table.)
Importing nodes and links
To create a network from one of the above mentioned data formats (adjacency matrix, link list, adjacency list) use the file menu, click on open..., select files of type CSV files (.txt, .csv) in the file chooser, navigate to the file you want to open, and click on the ok button. Then the import options dialog opens, where you select the appropriate data format in the topmost drop-down menu. Depending of the data format, the import options dialog shows you the various settings to be made, to interpret the data correctly. The image below shows the import options dialog, when selecting adjacency matrix as data format. The visualization and analysis tutorial explains the import of data from an adjacency matrix in detail, using an exemplary dataset. A description of importing all (other) data formats is given on the import options dialog page. Note that the import options dialog is designed to import information about nodes and links, not about attributes. The only exception is the import of node lists, where a network of isolated nodes and their attributes will be created.
Importing node and link attributes
To open an attribute table and add attribute values to the nodes or links of a network that is already opened in visone, use the attribute manager which can be started by clicking on the icon ![]() in visone's toolbar.
in visone's toolbar.
In the attribute manager, choose the node or link button in the top row, import & export on the left, operation import in the drop-down menu, and select the CSV file that contains the attributes you want to import
Before clicking on apply it is very important to correctly set the value in the join by drop-down menu. This should point to the name of the attribute that identifies the nodes (or links) and tells visone which column in the imported CSV file holds these identifiers. The identifying attribute must be identical to the header of the column that contains the identifiers (see below). The other columns contain the names and values of the other attributes. For instance, the node with id A gets a value of 23 for the age attribute and the value false for the smokes attribute. (When reading attribute tables you need column headers.)
id;age;smokes A;23;false B;28;true C;19;true D;27;false
Clicking on the apply button opens the import options which have the same meaning as when reading adjacency matrices (explained above).
Other supported formats
If nothing else works: use the R console or KNIME connection
visone offers the R console with which network data can be sent from visone to R and back. This ability to load data from R - together with R's data input and data processing capabilities - opens nearly unlimited possibilities to import data from yet other sources, as well as to import data that has to be cleaned, filtered, or preprocessed before turning it into a network.
The basic functioning of the visone to R interface is explained in the R tutorial. Tutorials about working with R are linked from the R website. A particular useful R package for data input is the foreign package for "reading and writing data stored by statistical packages such as Minitab, S, SAS, SPSS, Stata, Systat, ..., and for reading and writing dBase files."