R console (tutorial): Difference between revisions
m moved R-console (trail) to R console (trail) |
No edit summary |
||
| Line 1: | Line 1: | ||
This [[Trails|trail]] illustrates how to send network data from visone to R and back. [http://www.r-project.org/ The R project] for statistical computing offers a rich set of methods for data analysis and modeling which | This [[Trails|trail]] illustrates how to send network data from visone to R and back. [http://www.r-project.org/ The R project] for statistical computing offers a rich set of methods for data analysis and modeling which becomes accessible from visone through the R-console. We assume that you have installed the R connection as it is explained in the [[Installation_(trail)#Installing_the_R_connection|installation trail]]. This trail assumes that you have basic understanding about how to work with visone as it is, for instance, explained in the trail on [[Visualization_and_analysis_(trail)|visualization and analysis]]. You do ''not'' need to have any previous knowledge about R to follow this trail; nevertheless, to exploit the full potential of the R language you should consult the [http://www.r-project.org/ R-project page]. | ||
To follow the steps illustrated in this trail you should download the network file ''Egonet.graphml'' which is linked from and explained in the page [[Egoredes_(data)]]. Further you should remove all ties that are not rated as ''very likely'' in the same manner as it is explained in the last section of the [[Visualization_and_analysis_(trail)#Selecting_elements_dependent_on_attribute_values|visualization and analysis trail]]. | To follow the steps illustrated in this trail you should download the network file ''Egonet.graphml'' which is linked from and explained in the page [[Egoredes_(data)]]. Further you should remove all ties that are not rated as ''very likely'' in the same manner as it is explained in the last section of the [[Visualization_and_analysis_(trail)#Selecting_elements_dependent_on_attribute_values|visualization and analysis trail]]. | ||
Revision as of 18:20, 12 February 2011
This trail illustrates how to send network data from visone to R and back. The R project for statistical computing offers a rich set of methods for data analysis and modeling which becomes accessible from visone through the R-console. We assume that you have installed the R connection as it is explained in the installation trail. This trail assumes that you have basic understanding about how to work with visone as it is, for instance, explained in the trail on visualization and analysis. You do not need to have any previous knowledge about R to follow this trail; nevertheless, to exploit the full potential of the R language you should consult the R-project page.
To follow the steps illustrated in this trail you should download the network file Egonet.graphml which is linked from and explained in the page Egoredes_(data). Further you should remove all ties that are not rated as very likely in the same manner as it is explained in the last section of the visualization and analysis trail.
Sending networks from visone to R
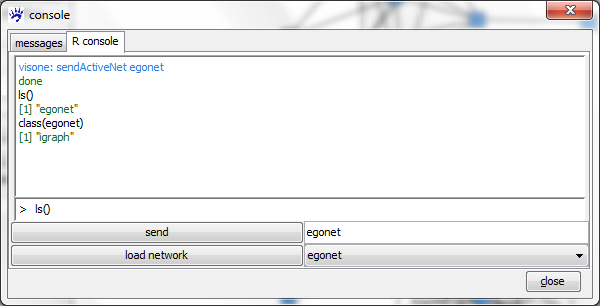
To send the network from visone to R open the R console tab, choose a name in the textfield r name (you might just accept visone's suggestion for this name, which should be egonet), and click on the send button.
After clicking on send visone starts the Rserve connection and opens the R-console. If this does not work you should check the settings of R connection options accessible from the file menu. If it works you should get a message like
ready to serve visone: sendActiveNet egonet done visone: lsIGraph "egonet"
in the message field of the R console.
You can list all variables that are in the R workspace by typing
ls()
in the input field at the bottom of the R console and pressing the Enter-Key (currently there is one object called egonet). As we'll see soon, egonet is an object of class igraph which is an R package obtainable from a CRAN Website (this site also gives you access to R tutorials and documentation). The igraph package is documented in more detail on http://igraph.sourceforge.net/.
Getting basic statistics about an igraph object
The class of the object egonet in the R workshop is printed when executing the command
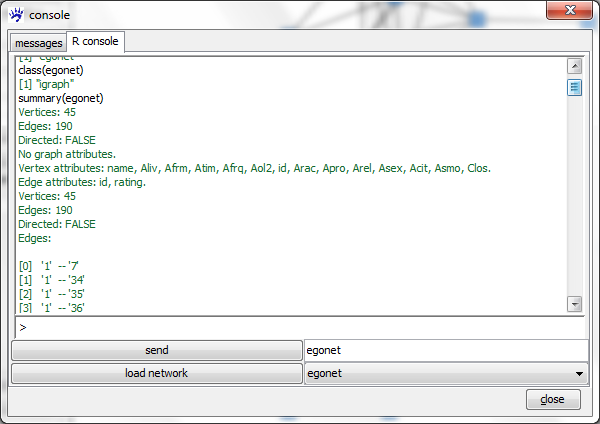
class(egonet)
As we can see the class of egonet is igraph. The igraph documentation linked above gives a complete list of all methods available for this class. In the following we describe how to inspect what is encoded in the given object and how to get simple summary statistics. Executing the command
summary(egonet)
outputs basic information such as the number of vertices and edges, the names of vertex and edge attributes, as well as a list of all edges.
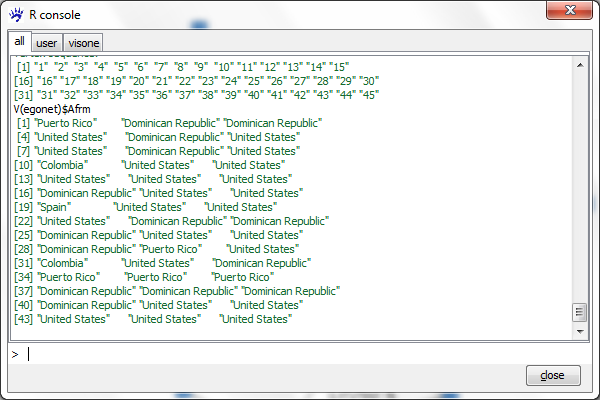
To see the values of the vertex or edge attributes one needs to understand the concept of vertex iterators and edge iterators in igraph. The vertex iterator of graph egonet is returned by typing the command
V(egonet)
When executing this you see just the list of vertex indices, i.e., the numbers from 1 to 45. The values of an attribute (e.g., Afrm) can be listed by typing
V(egonet)$Afrm