R console (tutorial)
This trail illustrates how to send network data from visone to R and back. The R project for statistical computing offers a rich set of methods for data analysis and modeling which becomes accessible from visone through the R console. We assume that you have installed the R connection as it is explained in the installation trail. This trail assumes that you have basic understanding about how to work with visone as it is, for instance, explained in the trail on visualization and analysis. You do not need to have any previous knowledge about R to follow this trail; nevertheless, to exploit the full potential offered by R you could consult documentation and tutorials linked from the R-project page.
To follow the steps illustrated in this trail you should download the network file Egonet.graphml which is linked from and explained in the page Egoredes_(data). Further you should remove all ties that are not rated as very likely in the same manner as it is explained in the last section of the visualization and analysis trail.
Sending networks from visone to R
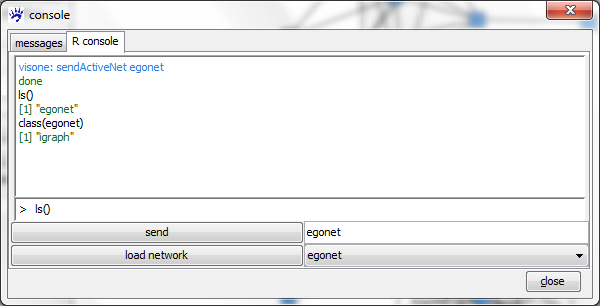
To send the network from visone to R open the R console tab, choose a name in the textfield r name (you might just accept visone's suggestion for this name, which should be egonet), and click on the send button. After clicking on send visone starts the Rserve connection and opens the R-console. If this does not work you should check the settings of the R connection options accessible via the file, options menu. If it works you should get a message like
ready to serve visone: sendActiveNet egonet done visone: lsIGraph "egonet"
in the message field of the R console.
You can list all variables that are in the R workspace by typing
ls()
in the input field at the bottom of the R console and pressing the Enter-Key (currently there is one object called egonet). As we'll see soon, egonet is an object of class igraph which is an R package obtainable from a CRAN Website (this site also gives you access to R tutorials and documentation). The igraph package is documented in more detail on http://igraph.sourceforge.net/.
Getting basic statistics about an igraph object
The class of the object egonet in the R workshop is printed when executing the command
class(egonet)
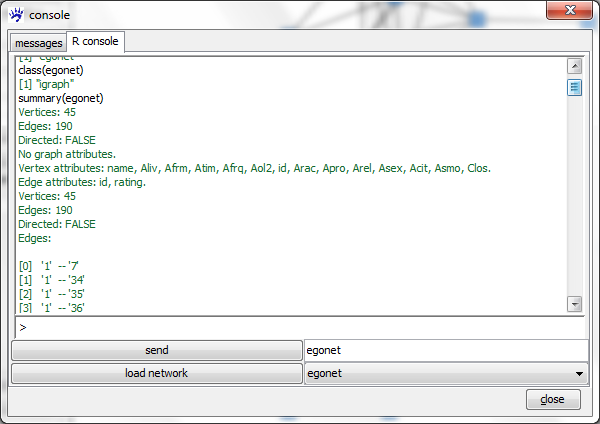
As we can see the class of egonet is igraph. The igraph documentation linked above gives a complete list of all methods available for this class. In the following we describe how to inspect what is encoded in the given object and how to get simple summary statistics. Executing the command
summary(egonet)
outputs basic information such as the number of vertices and edges, the names of vertex and edge attributes, as well as a list of all edges.
To see the values of the vertex or edge attributes one needs to understand the concept of vertex iterators and edge iterators in igraph. The vertex iterator of graph egonet is returned by typing the command
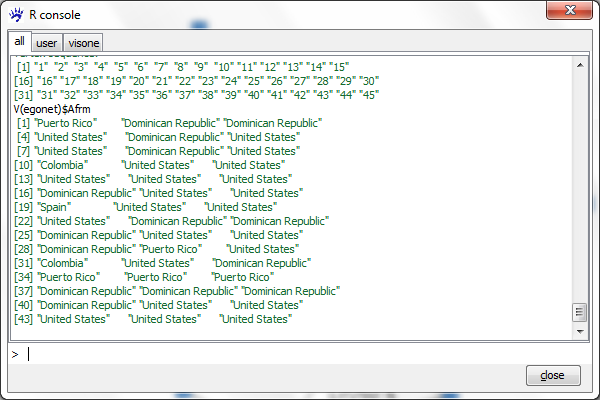
V(egonet)
When executing this you see just the list of vertex names which are here the numbers from 1 to 45. To obtain the values of an attribute (e.g., Afrm) type
V(egonet)$Afrm
which returns the vector of countries of origin of the various actors.
Useful summary statistics include information about how many actors originate from the various countries. However, typing the command
summary(V(egonet)$Afrm)
just outputs information about the class and size of the list of countries of origin:
Length Class Mode 45 character character
which is not very informative. To obtain the list of unique values for the Afrm attribute, you can type
unique(V(egonet)$Afrm)
which returns the names of five different countries. To count the number of actors in each of the countries, it is most convenient to convert the vector of character strings into a factor and save this factor in a new variable (e.g., called from) by typing the command
from <- as.factor(V(egonet)$Afrm)
Finally the command
summary(from)
returns the list of unique values along with the number of actors in each of the classes. In our example this is
Colombia Dominican Republic Puerto Rico Spain United States 2 14 5 1 23
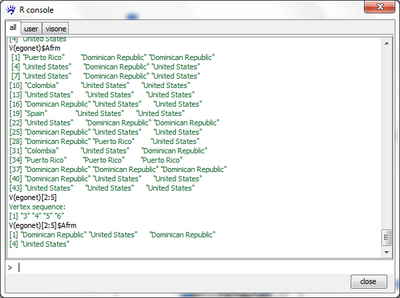
Vertex iterators can be restricted to subsets by specifying a logical vector (or a command that produces one) in square brackets after the iterator. For instance, the command
V(egonet)[2:5]
returns the values 3 to 6. This seeming contradiction is explained by the fact that counting of indices in vectors starts at zero; thus, the name of the vertex at position 0 is 1, the name of the vertex at position 2 is 3, and so on. The result of such a restriction operation on an vertex iterator is itself a vertex iterator and, thus, provides access to vertex attributes. For instance, typing
V(egonet)[2:5]$Afrm
returns the countries of origin of actors indexed by 2 to 5 (i.e., named 3 to 6). The command
V(egonet)[Acit == "new york"]$Afrm
gives you the countries of origin of all actors whose attribute Acit (encoding the city of residence) equals new york, and so on. Vertex iterators can be restricted by specifying more sophisticated conditions; see the igraph documentation for details.
An edge iterator is returned via the command
E(egonet)
and offers access to edge attributes similar as for vertices.
Analyzing distributions of centralities in igraph
The igraph package offers methods to compute various established centrality measures (refer to the igraph documentation for a complete list of available methods). While these could also be directly computed in visone without the detour via the R console, R directly offers statistical descriptions and analysis of the computed values. This is demonstrated in the following. The vertex degrees are returned by the command
degree(egonet)
Let's save this vector in a variable d by typing
d <- degree(egonet)
Mean, standard deviation, and summary statistics (including min, max, quartiles, and median) are computed by
mean(d) sd(d) summary(d)
To display these (or other) statistics separately for each class of actors defined by the country of origin (or any other attribute), execute the command
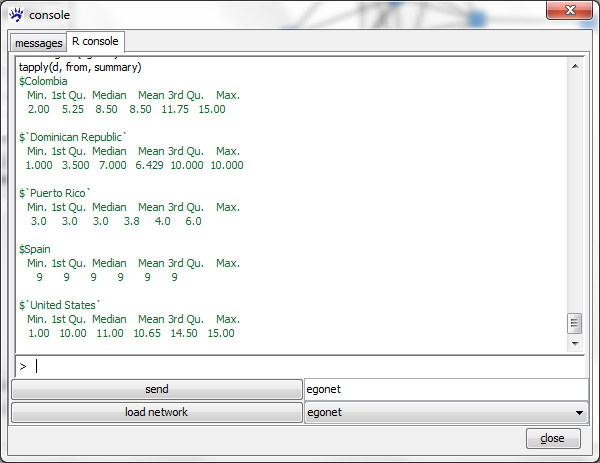
tapply(d, from, summary)
The three arguments of tapply have the following meaning: d is the vector of values to which the function should be applied, from is the factor whose unique values determine the different classes, and summary is the function to be computed (instead of summary, you could also type mean, sd, and so on).